---
layout: global
title: "Overview: estimators, transformers and pipelines - spark.ml"
displayTitle: "Overview: estimators, transformers and pipelines - spark.ml"
---
`\[
\newcommand{\R}{\mathbb{R}}
\newcommand{\E}{\mathbb{E}}
\newcommand{\x}{\mathbf{x}}
\newcommand{\y}{\mathbf{y}}
\newcommand{\wv}{\mathbf{w}}
\newcommand{\av}{\mathbf{\alpha}}
\newcommand{\bv}{\mathbf{b}}
\newcommand{\N}{\mathbb{N}}
\newcommand{\id}{\mathbf{I}}
\newcommand{\ind}{\mathbf{1}}
\newcommand{\0}{\mathbf{0}}
\newcommand{\unit}{\mathbf{e}}
\newcommand{\one}{\mathbf{1}}
\newcommand{\zero}{\mathbf{0}}
\]`
The `spark.ml` package aims to provide a uniform set of high-level APIs built on top of
[DataFrames](sql-programming-guide.html#dataframes) that help users create and tune practical
machine learning pipelines.
See the [algorithm guides](#algorithm-guides) section below for guides on sub-packages of
`spark.ml`, including feature transformers unique to the Pipelines API, ensembles, and more.
**Table of contents**
* This will become a table of contents (this text will be scraped).
{:toc}
# Main concepts in Pipelines
Spark ML standardizes APIs for machine learning algorithms to make it easier to combine multiple
algorithms into a single pipeline, or workflow.
This section covers the key concepts introduced by the Spark ML API, where the pipeline concept is
mostly inspired by the [scikit-learn](http://scikit-learn.org/) project.
* **[`DataFrame`](ml-guide.html#dataframe)**: Spark ML uses `DataFrame` from Spark SQL as an ML
dataset, which can hold a variety of data types.
E.g., a `DataFrame` could have different columns storing text, feature vectors, true labels, and predictions.
* **[`Transformer`](ml-guide.html#transformers)**: A `Transformer` is an algorithm which can transform one `DataFrame` into another `DataFrame`.
E.g., an ML model is a `Transformer` which transforms `DataFrame` with features into a `DataFrame` with predictions.
* **[`Estimator`](ml-guide.html#estimators)**: An `Estimator` is an algorithm which can be fit on a `DataFrame` to produce a `Transformer`.
E.g., a learning algorithm is an `Estimator` which trains on a `DataFrame` and produces a model.
* **[`Pipeline`](ml-guide.html#pipeline)**: A `Pipeline` chains multiple `Transformer`s and `Estimator`s together to specify an ML workflow.
* **[`Parameter`](ml-guide.html#parameters)**: All `Transformer`s and `Estimator`s now share a common API for specifying parameters.
## DataFrame
Machine learning can be applied to a wide variety of data types, such as vectors, text, images, and structured data.
Spark ML adopts the `DataFrame` from Spark SQL in order to support a variety of data types.
`DataFrame` supports many basic and structured types; see the [Spark SQL datatype reference](sql-programming-guide.html#spark-sql-datatype-reference) for a list of supported types.
In addition to the types listed in the Spark SQL guide, `DataFrame` can use ML [`Vector`](mllib-data-types.html#local-vector) types.
A `DataFrame` can be created either implicitly or explicitly from a regular `RDD`. See the code examples below and the [Spark SQL programming guide](sql-programming-guide.html) for examples.
Columns in a `DataFrame` are named. The code examples below use names such as "text," "features," and "label."
## Pipeline components
### Transformers
A `Transformer` is an abstraction that includes feature transformers and learned models.
Technically, a `Transformer` implements a method `transform()`, which converts one `DataFrame` into
another, generally by appending one or more columns.
For example:
* A feature transformer might take a `DataFrame`, read a column (e.g., text), map it into a new
column (e.g., feature vectors), and output a new `DataFrame` with the mapped column appended.
* A learning model might take a `DataFrame`, read the column containing feature vectors, predict the
label for each feature vector, and output a new `DataFrame` with predicted labels appended as a
column.
### Estimators
An `Estimator` abstracts the concept of a learning algorithm or any algorithm that fits or trains on
data.
Technically, an `Estimator` implements a method `fit()`, which accepts a `DataFrame` and produces a
`Model`, which is a `Transformer`.
For example, a learning algorithm such as `LogisticRegression` is an `Estimator`, and calling
`fit()` trains a `LogisticRegressionModel`, which is a `Model` and hence a `Transformer`.
### Properties of pipeline components
`Transformer.transform()`s and `Estimator.fit()`s are both stateless. In the future, stateful algorithms may be supported via alternative concepts.
Each instance of a `Transformer` or `Estimator` has a unique ID, which is useful in specifying parameters (discussed below).
## Pipeline
In machine learning, it is common to run a sequence of algorithms to process and learn from data.
E.g., a simple text document processing workflow might include several stages:
* Split each document's text into words.
* Convert each document's words into a numerical feature vector.
* Learn a prediction model using the feature vectors and labels.
Spark ML represents such a workflow as a `Pipeline`, which consists of a sequence of
`PipelineStage`s (`Transformer`s and `Estimator`s) to be run in a specific order.
We will use this simple workflow as a running example in this section.
### How it works
A `Pipeline` is specified as a sequence of stages, and each stage is either a `Transformer` or an `Estimator`.
These stages are run in order, and the input `DataFrame` is transformed as it passes through each stage.
For `Transformer` stages, the `transform()` method is called on the `DataFrame`.
For `Estimator` stages, the `fit()` method is called to produce a `Transformer` (which becomes part of the `PipelineModel`, or fitted `Pipeline`), and that `Transformer`'s `transform()` method is called on the `DataFrame`.
We illustrate this for the simple text document workflow. The figure below is for the *training time* usage of a `Pipeline`.
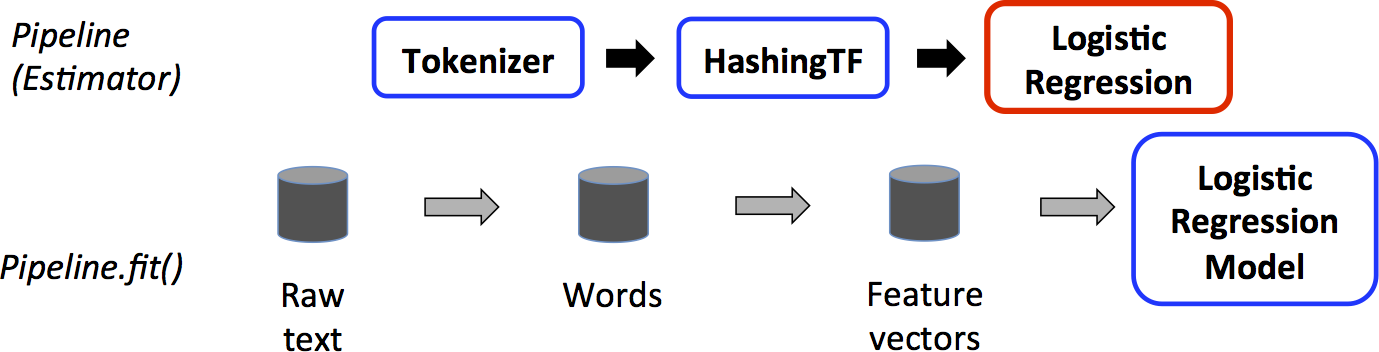
Above, the top row represents a `Pipeline` with three stages.
The first two (`Tokenizer` and `HashingTF`) are `Transformer`s (blue), and the third (`LogisticRegression`) is an `Estimator` (red).
The bottom row represents data flowing through the pipeline, where cylinders indicate `DataFrame`s.
The `Pipeline.fit()` method is called on the original `DataFrame`, which has raw text documents and labels.
The `Tokenizer.transform()` method splits the raw text documents into words, adding a new column with words to the `DataFrame`.
The `HashingTF.transform()` method converts the words column into feature vectors, adding a new column with those vectors to the `DataFrame`.
Now, since `LogisticRegression` is an `Estimator`, the `Pipeline` first calls `LogisticRegression.fit()` to produce a `LogisticRegressionModel`.
If the `Pipeline` had more stages, it would call the `LogisticRegressionModel`'s `transform()`
method on the `DataFrame` before passing the `DataFrame` to the next stage.
A `Pipeline` is an `Estimator`.
Thus, after a `Pipeline`'s `fit()` method runs, it produces a `PipelineModel`, which is a
`Transformer`.
This `PipelineModel` is used at *test time*; the figure below illustrates this usage.
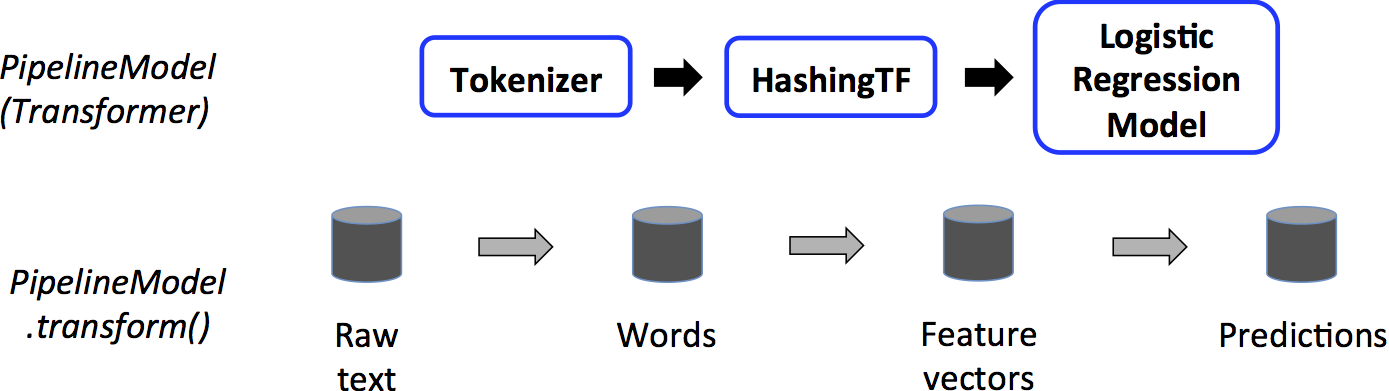
In the figure above, the `PipelineModel` has the same number of stages as the original `Pipeline`, but all `Estimator`s in the original `Pipeline` have become `Transformer`s.
When the `PipelineModel`'s `transform()` method is called on a test dataset, the data are passed
through the fitted pipeline in order.
Each stage's `transform()` method updates the dataset and passes it to the next stage.
`Pipeline`s and `PipelineModel`s help to ensure that training and test data go through identical feature processing steps.
### Details
*DAG `Pipeline`s*: A `Pipeline`'s stages are specified as an ordered array. The examples given here are all for linear `Pipeline`s, i.e., `Pipeline`s in which each stage uses data produced by the previous stage. It is possible to create non-linear `Pipeline`s as long as the data flow graph forms a Directed Acyclic Graph (DAG). This graph is currently specified implicitly based on the input and output column names of each stage (generally specified as parameters). If the `Pipeline` forms a DAG, then the stages must be specified in topological order.
*Runtime checking*: Since `Pipeline`s can operate on `DataFrame`s with varied types, they cannot use
compile-time type checking.
`Pipeline`s and `PipelineModel`s instead do runtime checking before actually running the `Pipeline`.
This type checking is done using the `DataFrame` *schema*, a description of the data types of columns in the `DataFrame`.
*Unique Pipeline stages*: A `Pipeline`'s stages should be unique instances. E.g., the same instance
`myHashingTF` should not be inserted into the `Pipeline` twice since `Pipeline` stages must have
unique IDs. However, different instances `myHashingTF1` and `myHashingTF2` (both of type `HashingTF`)
can be put into the same `Pipeline` since different instances will be created with different IDs.
## Parameters
Spark ML `Estimator`s and `Transformer`s use a uniform API for specifying parameters.
A `Param` is a named parameter with self-contained documentation.
A `ParamMap` is a set of (parameter, value) pairs.
There are two main ways to pass parameters to an algorithm:
1. Set parameters for an instance. E.g., if `lr` is an instance of `LogisticRegression`, one could
call `lr.setMaxIter(10)` to make `lr.fit()` use at most 10 iterations.
This API resembles the API used in `spark.mllib` package.
2. Pass a `ParamMap` to `fit()` or `transform()`. Any parameters in the `ParamMap` will override parameters previously specified via setter methods.
Parameters belong to specific instances of `Estimator`s and `Transformer`s.
For example, if we have two `LogisticRegression` instances `lr1` and `lr2`, then we can build a `ParamMap` with both `maxIter` parameters specified: `ParamMap(lr1.maxIter -> 10, lr2.maxIter -> 20)`.
This is useful if there are two algorithms with the `maxIter` parameter in a `Pipeline`.
## Saving and Loading Pipelines
Often times it is worth it to save a model or a pipeline to disk for later use. In Spark 1.6, a model import/export functionality was added to the Pipeline API. Most basic transformers are supported as well as some of the more basic ML models. Please refer to the algorithm's API documentation to see if saving and loading is supported.
# Code examples
This section gives code examples illustrating the functionality discussed above.
For more info, please refer to the API documentation
([Scala](api/scala/index.html#org.apache.spark.ml.package),
[Java](api/java/org/apache/spark/ml/package-summary.html),
and [Python](api/python/pyspark.ml.html)).
Some Spark ML algorithms are wrappers for `spark.mllib` algorithms, and the
[MLlib programming guide](mllib-guide.html) has details on specific algorithms.
## Example: Estimator, Transformer, and Param
This example covers the concepts of `Estimator`, `Transformer`, and `Param`.
{% highlight scala %}
import org.apache.spark.ml.classification.LogisticRegression
import org.apache.spark.ml.param.ParamMap
import org.apache.spark.mllib.linalg.{Vector, Vectors}
import org.apache.spark.sql.Row
// Prepare training data from a list of (label, features) tuples.
val training = sqlContext.createDataFrame(Seq(
(1.0, Vectors.dense(0.0, 1.1, 0.1)),
(0.0, Vectors.dense(2.0, 1.0, -1.0)),
(0.0, Vectors.dense(2.0, 1.3, 1.0)),
(1.0, Vectors.dense(0.0, 1.2, -0.5))
)).toDF("label", "features")
// Create a LogisticRegression instance. This instance is an Estimator.
val lr = new LogisticRegression()
// Print out the parameters, documentation, and any default values.
println("LogisticRegression parameters:\n" + lr.explainParams() + "\n")
// We may set parameters using setter methods.
lr.setMaxIter(10)
.setRegParam(0.01)
// Learn a LogisticRegression model. This uses the parameters stored in lr.
val model1 = lr.fit(training)
// Since model1 is a Model (i.e., a Transformer produced by an Estimator),
// we can view the parameters it used during fit().
// This prints the parameter (name: value) pairs, where names are unique IDs for this
// LogisticRegression instance.
println("Model 1 was fit using parameters: " + model1.parent.extractParamMap)
// We may alternatively specify parameters using a ParamMap,
// which supports several methods for specifying parameters.
val paramMap = ParamMap(lr.maxIter -> 20)
.put(lr.maxIter, 30) // Specify 1 Param. This overwrites the original maxIter.
.put(lr.regParam -> 0.1, lr.threshold -> 0.55) // Specify multiple Params.
// One can also combine ParamMaps.
val paramMap2 = ParamMap(lr.probabilityCol -> "myProbability") // Change output column name
val paramMapCombined = paramMap ++ paramMap2
// Now learn a new model using the paramMapCombined parameters.
// paramMapCombined overrides all parameters set earlier via lr.set* methods.
val model2 = lr.fit(training, paramMapCombined)
println("Model 2 was fit using parameters: " + model2.parent.extractParamMap)
// Prepare test data.
val test = sqlContext.createDataFrame(Seq(
(1.0, Vectors.dense(-1.0, 1.5, 1.3)),
(0.0, Vectors.dense(3.0, 2.0, -0.1)),
(1.0, Vectors.dense(0.0, 2.2, -1.5))
)).toDF("label", "features")
// Make predictions on test data using the Transformer.transform() method.
// LogisticRegression.transform will only use the 'features' column.
// Note that model2.transform() outputs a 'myProbability' column instead of the usual
// 'probability' column since we renamed the lr.probabilityCol parameter previously.
model2.transform(test)
.select("features", "label", "myProbability", "prediction")
.collect()
.foreach { case Row(features: Vector, label: Double, prob: Vector, prediction: Double) =>
println(s"($features, $label) -> prob=$prob, prediction=$prediction")
}
{% endhighlight %}
{% highlight java %}
import java.util.Arrays;
import java.util.List;
import org.apache.spark.ml.classification.LogisticRegressionModel;
import org.apache.spark.ml.param.ParamMap;
import org.apache.spark.ml.classification.LogisticRegression;
import org.apache.spark.mllib.linalg.Vectors;
import org.apache.spark.mllib.regression.LabeledPoint;
import org.apache.spark.sql.DataFrame;
import org.apache.spark.sql.Row;
// Prepare training data.
// We use LabeledPoint, which is a JavaBean. Spark SQL can convert RDDs of JavaBeans
// into DataFrames, where it uses the bean metadata to infer the schema.
DataFrame training = sqlContext.createDataFrame(Arrays.asList(
new LabeledPoint(1.0, Vectors.dense(0.0, 1.1, 0.1)),
new LabeledPoint(0.0, Vectors.dense(2.0, 1.0, -1.0)),
new LabeledPoint(0.0, Vectors.dense(2.0, 1.3, 1.0)),
new LabeledPoint(1.0, Vectors.dense(0.0, 1.2, -0.5))
), LabeledPoint.class);
// Create a LogisticRegression instance. This instance is an Estimator.
LogisticRegression lr = new LogisticRegression();
// Print out the parameters, documentation, and any default values.
System.out.println("LogisticRegression parameters:\n" + lr.explainParams() + "\n");
// We may set parameters using setter methods.
lr.setMaxIter(10)
.setRegParam(0.01);
// Learn a LogisticRegression model. This uses the parameters stored in lr.
LogisticRegressionModel model1 = lr.fit(training);
// Since model1 is a Model (i.e., a Transformer produced by an Estimator),
// we can view the parameters it used during fit().
// This prints the parameter (name: value) pairs, where names are unique IDs for this
// LogisticRegression instance.
System.out.println("Model 1 was fit using parameters: " + model1.parent().extractParamMap());
// We may alternatively specify parameters using a ParamMap.
ParamMap paramMap = new ParamMap()
.put(lr.maxIter().w(20)) // Specify 1 Param.
.put(lr.maxIter(), 30) // This overwrites the original maxIter.
.put(lr.regParam().w(0.1), lr.threshold().w(0.55)); // Specify multiple Params.
// One can also combine ParamMaps.
ParamMap paramMap2 = new ParamMap()
.put(lr.probabilityCol().w("myProbability")); // Change output column name
ParamMap paramMapCombined = paramMap.$plus$plus(paramMap2);
// Now learn a new model using the paramMapCombined parameters.
// paramMapCombined overrides all parameters set earlier via lr.set* methods.
LogisticRegressionModel model2 = lr.fit(training, paramMapCombined);
System.out.println("Model 2 was fit using parameters: " + model2.parent().extractParamMap());
// Prepare test documents.
DataFrame test = sqlContext.createDataFrame(Arrays.asList(
new LabeledPoint(1.0, Vectors.dense(-1.0, 1.5, 1.3)),
new LabeledPoint(0.0, Vectors.dense(3.0, 2.0, -0.1)),
new LabeledPoint(1.0, Vectors.dense(0.0, 2.2, -1.5))
), LabeledPoint.class);
// Make predictions on test documents using the Transformer.transform() method.
// LogisticRegression.transform will only use the 'features' column.
// Note that model2.transform() outputs a 'myProbability' column instead of the usual
// 'probability' column since we renamed the lr.probabilityCol parameter previously.
DataFrame results = model2.transform(test);
for (Row r: results.select("features", "label", "myProbability", "prediction").collect()) {
System.out.println("(" + r.get(0) + ", " + r.get(1) + ") -> prob=" + r.get(2)
+ ", prediction=" + r.get(3));
}
{% endhighlight %}
{% highlight python %}
from pyspark.mllib.linalg import Vectors
from pyspark.ml.classification import LogisticRegression
from pyspark.ml.param import Param, Params
# Prepare training data from a list of (label, features) tuples.
training = sqlContext.createDataFrame([
(1.0, Vectors.dense([0.0, 1.1, 0.1])),
(0.0, Vectors.dense([2.0, 1.0, -1.0])),
(0.0, Vectors.dense([2.0, 1.3, 1.0])),
(1.0, Vectors.dense([0.0, 1.2, -0.5]))], ["label", "features"])
# Create a LogisticRegression instance. This instance is an Estimator.
lr = LogisticRegression(maxIter=10, regParam=0.01)
# Print out the parameters, documentation, and any default values.
print "LogisticRegression parameters:\n" + lr.explainParams() + "\n"
# Learn a LogisticRegression model. This uses the parameters stored in lr.
model1 = lr.fit(training)
# Since model1 is a Model (i.e., a transformer produced by an Estimator),
# we can view the parameters it used during fit().
# This prints the parameter (name: value) pairs, where names are unique IDs for this
# LogisticRegression instance.
print "Model 1 was fit using parameters: "
print model1.extractParamMap()
# We may alternatively specify parameters using a Python dictionary as a paramMap
paramMap = {lr.maxIter: 20}
paramMap[lr.maxIter] = 30 # Specify 1 Param, overwriting the original maxIter.
paramMap.update({lr.regParam: 0.1, lr.threshold: 0.55}) # Specify multiple Params.
# You can combine paramMaps, which are python dictionaries.
paramMap2 = {lr.probabilityCol: "myProbability"} # Change output column name
paramMapCombined = paramMap.copy()
paramMapCombined.update(paramMap2)
# Now learn a new model using the paramMapCombined parameters.
# paramMapCombined overrides all parameters set earlier via lr.set* methods.
model2 = lr.fit(training, paramMapCombined)
print "Model 2 was fit using parameters: "
print model2.extractParamMap()
# Prepare test data
test = sqlContext.createDataFrame([
(1.0, Vectors.dense([-1.0, 1.5, 1.3])),
(0.0, Vectors.dense([3.0, 2.0, -0.1])),
(1.0, Vectors.dense([0.0, 2.2, -1.5]))], ["label", "features"])
# Make predictions on test data using the Transformer.transform() method.
# LogisticRegression.transform will only use the 'features' column.
# Note that model2.transform() outputs a "myProbability" column instead of the usual
# 'probability' column since we renamed the lr.probabilityCol parameter previously.
prediction = model2.transform(test)
selected = prediction.select("features", "label", "myProbability", "prediction")
for row in selected.collect():
print row
{% endhighlight %}
## Example: Pipeline
This example follows the simple text document `Pipeline` illustrated in the figures above.
{% highlight scala %}
import org.apache.spark.ml.{Pipeline, PipelineModel}
import org.apache.spark.ml.classification.LogisticRegression
import org.apache.spark.ml.feature.{HashingTF, Tokenizer}
import org.apache.spark.mllib.linalg.Vector
import org.apache.spark.sql.Row
// Prepare training documents from a list of (id, text, label) tuples.
val training = sqlContext.createDataFrame(Seq(
(0L, "a b c d e spark", 1.0),
(1L, "b d", 0.0),
(2L, "spark f g h", 1.0),
(3L, "hadoop mapreduce", 0.0)
)).toDF("id", "text", "label")
// Configure an ML pipeline, which consists of three stages: tokenizer, hashingTF, and lr.
val tokenizer = new Tokenizer()
.setInputCol("text")
.setOutputCol("words")
val hashingTF = new HashingTF()
.setNumFeatures(1000)
.setInputCol(tokenizer.getOutputCol)
.setOutputCol("features")
val lr = new LogisticRegression()
.setMaxIter(10)
.setRegParam(0.01)
val pipeline = new Pipeline()
.setStages(Array(tokenizer, hashingTF, lr))
// Fit the pipeline to training documents.
val model = pipeline.fit(training)
// now we can optionally save the fitted pipeline to disk
model.save("/tmp/spark-logistic-regression-model")
// we can also save this unfit pipeline to disk
pipeline.save("/tmp/unfit-lr-model")
// and load it back in during production
val sameModel = PipelineModel.load("/tmp/spark-logistic-regression-model")
// Prepare test documents, which are unlabeled (id, text) tuples.
val test = sqlContext.createDataFrame(Seq(
(4L, "spark i j k"),
(5L, "l m n"),
(6L, "mapreduce spark"),
(7L, "apache hadoop")
)).toDF("id", "text")
// Make predictions on test documents.
model.transform(test)
.select("id", "text", "probability", "prediction")
.collect()
.foreach { case Row(id: Long, text: String, prob: Vector, prediction: Double) =>
println(s"($id, $text) --> prob=$prob, prediction=$prediction")
}
{% endhighlight %}
{% highlight java %}
import java.util.Arrays;
import java.util.List;
import org.apache.spark.ml.Pipeline;
import org.apache.spark.ml.PipelineModel;
import org.apache.spark.ml.PipelineStage;
import org.apache.spark.ml.classification.LogisticRegression;
import org.apache.spark.ml.feature.HashingTF;
import org.apache.spark.ml.feature.Tokenizer;
import org.apache.spark.sql.DataFrame;
import org.apache.spark.sql.Row;
// Labeled and unlabeled instance types.
// Spark SQL can infer schema from Java Beans.
public class Document implements Serializable {
private long id;
private String text;
public Document(long id, String text) {
this.id = id;
this.text = text;
}
public long getId() { return this.id; }
public void setId(long id) { this.id = id; }
public String getText() { return this.text; }
public void setText(String text) { this.text = text; }
}
public class LabeledDocument extends Document implements Serializable {
private double label;
public LabeledDocument(long id, String text, double label) {
super(id, text);
this.label = label;
}
public double getLabel() { return this.label; }
public void setLabel(double label) { this.label = label; }
}
// Prepare training documents, which are labeled.
DataFrame training = sqlContext.createDataFrame(Arrays.asList(
new LabeledDocument(0L, "a b c d e spark", 1.0),
new LabeledDocument(1L, "b d", 0.0),
new LabeledDocument(2L, "spark f g h", 1.0),
new LabeledDocument(3L, "hadoop mapreduce", 0.0)
), LabeledDocument.class);
// Configure an ML pipeline, which consists of three stages: tokenizer, hashingTF, and lr.
Tokenizer tokenizer = new Tokenizer()
.setInputCol("text")
.setOutputCol("words");
HashingTF hashingTF = new HashingTF()
.setNumFeatures(1000)
.setInputCol(tokenizer.getOutputCol())
.setOutputCol("features");
LogisticRegression lr = new LogisticRegression()
.setMaxIter(10)
.setRegParam(0.01);
Pipeline pipeline = new Pipeline()
.setStages(new PipelineStage[] {tokenizer, hashingTF, lr});
// Fit the pipeline to training documents.
PipelineModel model = pipeline.fit(training);
// Prepare test documents, which are unlabeled.
DataFrame test = sqlContext.createDataFrame(Arrays.asList(
new Document(4L, "spark i j k"),
new Document(5L, "l m n"),
new Document(6L, "mapreduce spark"),
new Document(7L, "apache hadoop")
), Document.class);
// Make predictions on test documents.
DataFrame predictions = model.transform(test);
for (Row r: predictions.select("id", "text", "probability", "prediction").collect()) {
System.out.println("(" + r.get(0) + ", " + r.get(1) + ") --> prob=" + r.get(2)
+ ", prediction=" + r.get(3));
}
{% endhighlight %}
{% highlight python %}
from pyspark.ml import Pipeline
from pyspark.ml.classification import LogisticRegression
from pyspark.ml.feature import HashingTF, Tokenizer
from pyspark.sql import Row
# Prepare training documents from a list of (id, text, label) tuples.
LabeledDocument = Row("id", "text", "label")
training = sqlContext.createDataFrame([
(0L, "a b c d e spark", 1.0),
(1L, "b d", 0.0),
(2L, "spark f g h", 1.0),
(3L, "hadoop mapreduce", 0.0)], ["id", "text", "label"])
# Configure an ML pipeline, which consists of tree stages: tokenizer, hashingTF, and lr.
tokenizer = Tokenizer(inputCol="text", outputCol="words")
hashingTF = HashingTF(inputCol=tokenizer.getOutputCol(), outputCol="features")
lr = LogisticRegression(maxIter=10, regParam=0.01)
pipeline = Pipeline(stages=[tokenizer, hashingTF, lr])
# Fit the pipeline to training documents.
model = pipeline.fit(training)
# Prepare test documents, which are unlabeled (id, text) tuples.
test = sqlContext.createDataFrame([
(4L, "spark i j k"),
(5L, "l m n"),
(6L, "mapreduce spark"),
(7L, "apache hadoop")], ["id", "text"])
# Make predictions on test documents and print columns of interest.
prediction = model.transform(test)
selected = prediction.select("id", "text", "prediction")
for row in selected.collect():
print(row)
{% endhighlight %}
## Example: model selection via cross-validation
An important task in ML is *model selection*, or using data to find the best model or parameters for a given task. This is also called *tuning*.
`Pipeline`s facilitate model selection by making it easy to tune an entire `Pipeline` at once, rather than tuning each element in the `Pipeline` separately.
Currently, `spark.ml` supports model selection using the [`CrossValidator`](api/scala/index.html#org.apache.spark.ml.tuning.CrossValidator) class, which takes an `Estimator`, a set of `ParamMap`s, and an [`Evaluator`](api/scala/index.html#org.apache.spark.ml.evaluation.Evaluator).
`CrossValidator` begins by splitting the dataset into a set of *folds* which are used as separate training and test datasets; e.g., with `$k=3$` folds, `CrossValidator` will generate 3 (training, test) dataset pairs, each of which uses 2/3 of the data for training and 1/3 for testing.
`CrossValidator` iterates through the set of `ParamMap`s. For each `ParamMap`, it trains the given `Estimator` and evaluates it using the given `Evaluator`.
The `Evaluator` can be a [`RegressionEvaluator`](api/scala/index.html#org.apache.spark.ml.evaluation.RegressionEvaluator)
for regression problems, a [`BinaryClassificationEvaluator`](api/scala/index.html#org.apache.spark.ml.evaluation.BinaryClassificationEvaluator)
for binary data, or a [`MultiClassClassificationEvaluator`](api/scala/index.html#org.apache.spark.ml.evaluation.MulticlassClassificationEvaluator)
for multiclass problems. The default metric used to choose the best `ParamMap` can be overriden by the `setMetricName`
method in each of these evaluators.
The `ParamMap` which produces the best evaluation metric (averaged over the `$k$` folds) is selected as the best model.
`CrossValidator` finally fits the `Estimator` using the best `ParamMap` and the entire dataset.
The following example demonstrates using `CrossValidator` to select from a grid of parameters.
To help construct the parameter grid, we use the [`ParamGridBuilder`](api/scala/index.html#org.apache.spark.ml.tuning.ParamGridBuilder) utility.
Note that cross-validation over a grid of parameters is expensive.
E.g., in the example below, the parameter grid has 3 values for `hashingTF.numFeatures` and 2 values for `lr.regParam`, and `CrossValidator` uses 2 folds. This multiplies out to `$(3 \times 2) \times 2 = 12$` different models being trained.
In realistic settings, it can be common to try many more parameters and use more folds (`$k=3$` and `$k=10$` are common).
In other words, using `CrossValidator` can be very expensive.
However, it is also a well-established method for choosing parameters which is more statistically sound than heuristic hand-tuning.
{% highlight scala %}
import org.apache.spark.ml.Pipeline
import org.apache.spark.ml.classification.LogisticRegression
import org.apache.spark.ml.evaluation.BinaryClassificationEvaluator
import org.apache.spark.ml.feature.{HashingTF, Tokenizer}
import org.apache.spark.ml.tuning.{ParamGridBuilder, CrossValidator}
import org.apache.spark.mllib.linalg.Vector
import org.apache.spark.sql.Row
// Prepare training data from a list of (id, text, label) tuples.
val training = sqlContext.createDataFrame(Seq(
(0L, "a b c d e spark", 1.0),
(1L, "b d", 0.0),
(2L, "spark f g h", 1.0),
(3L, "hadoop mapreduce", 0.0),
(4L, "b spark who", 1.0),
(5L, "g d a y", 0.0),
(6L, "spark fly", 1.0),
(7L, "was mapreduce", 0.0),
(8L, "e spark program", 1.0),
(9L, "a e c l", 0.0),
(10L, "spark compile", 1.0),
(11L, "hadoop software", 0.0)
)).toDF("id", "text", "label")
// Configure an ML pipeline, which consists of three stages: tokenizer, hashingTF, and lr.
val tokenizer = new Tokenizer()
.setInputCol("text")
.setOutputCol("words")
val hashingTF = new HashingTF()
.setInputCol(tokenizer.getOutputCol)
.setOutputCol("features")
val lr = new LogisticRegression()
.setMaxIter(10)
val pipeline = new Pipeline()
.setStages(Array(tokenizer, hashingTF, lr))
// We use a ParamGridBuilder to construct a grid of parameters to search over.
// With 3 values for hashingTF.numFeatures and 2 values for lr.regParam,
// this grid will have 3 x 2 = 6 parameter settings for CrossValidator to choose from.
val paramGrid = new ParamGridBuilder()
.addGrid(hashingTF.numFeatures, Array(10, 100, 1000))
.addGrid(lr.regParam, Array(0.1, 0.01))
.build()
// We now treat the Pipeline as an Estimator, wrapping it in a CrossValidator instance.
// This will allow us to jointly choose parameters for all Pipeline stages.
// A CrossValidator requires an Estimator, a set of Estimator ParamMaps, and an Evaluator.
// Note that the evaluator here is a BinaryClassificationEvaluator and its default metric
// is areaUnderROC.
val cv = new CrossValidator()
.setEstimator(pipeline)
.setEvaluator(new BinaryClassificationEvaluator)
.setEstimatorParamMaps(paramGrid)
.setNumFolds(2) // Use 3+ in practice
// Run cross-validation, and choose the best set of parameters.
val cvModel = cv.fit(training)
// Prepare test documents, which are unlabeled (id, text) tuples.
val test = sqlContext.createDataFrame(Seq(
(4L, "spark i j k"),
(5L, "l m n"),
(6L, "mapreduce spark"),
(7L, "apache hadoop")
)).toDF("id", "text")
// Make predictions on test documents. cvModel uses the best model found (lrModel).
cvModel.transform(test)
.select("id", "text", "probability", "prediction")
.collect()
.foreach { case Row(id: Long, text: String, prob: Vector, prediction: Double) =>
println(s"($id, $text) --> prob=$prob, prediction=$prediction")
}
{% endhighlight %}
{% highlight java %}
import java.util.Arrays;
import java.util.List;
import org.apache.spark.ml.Pipeline;
import org.apache.spark.ml.PipelineStage;
import org.apache.spark.ml.classification.LogisticRegression;
import org.apache.spark.ml.evaluation.BinaryClassificationEvaluator;
import org.apache.spark.ml.feature.HashingTF;
import org.apache.spark.ml.feature.Tokenizer;
import org.apache.spark.ml.param.ParamMap;
import org.apache.spark.ml.tuning.CrossValidator;
import org.apache.spark.ml.tuning.CrossValidatorModel;
import org.apache.spark.ml.tuning.ParamGridBuilder;
import org.apache.spark.sql.DataFrame;
import org.apache.spark.sql.Row;
// Labeled and unlabeled instance types.
// Spark SQL can infer schema from Java Beans.
public class Document implements Serializable {
private long id;
private String text;
public Document(long id, String text) {
this.id = id;
this.text = text;
}
public long getId() { return this.id; }
public void setId(long id) { this.id = id; }
public String getText() { return this.text; }
public void setText(String text) { this.text = text; }
}
public class LabeledDocument extends Document implements Serializable {
private double label;
public LabeledDocument(long id, String text, double label) {
super(id, text);
this.label = label;
}
public double getLabel() { return this.label; }
public void setLabel(double label) { this.label = label; }
}
// Prepare training documents, which are labeled.
DataFrame training = sqlContext.createDataFrame(Arrays.asList(
new LabeledDocument(0L, "a b c d e spark", 1.0),
new LabeledDocument(1L, "b d", 0.0),
new LabeledDocument(2L, "spark f g h", 1.0),
new LabeledDocument(3L, "hadoop mapreduce", 0.0),
new LabeledDocument(4L, "b spark who", 1.0),
new LabeledDocument(5L, "g d a y", 0.0),
new LabeledDocument(6L, "spark fly", 1.0),
new LabeledDocument(7L, "was mapreduce", 0.0),
new LabeledDocument(8L, "e spark program", 1.0),
new LabeledDocument(9L, "a e c l", 0.0),
new LabeledDocument(10L, "spark compile", 1.0),
new LabeledDocument(11L, "hadoop software", 0.0)
), LabeledDocument.class);
// Configure an ML pipeline, which consists of three stages: tokenizer, hashingTF, and lr.
Tokenizer tokenizer = new Tokenizer()
.setInputCol("text")
.setOutputCol("words");
HashingTF hashingTF = new HashingTF()
.setNumFeatures(1000)
.setInputCol(tokenizer.getOutputCol())
.setOutputCol("features");
LogisticRegression lr = new LogisticRegression()
.setMaxIter(10)
.setRegParam(0.01);
Pipeline pipeline = new Pipeline()
.setStages(new PipelineStage[] {tokenizer, hashingTF, lr});
// We use a ParamGridBuilder to construct a grid of parameters to search over.
// With 3 values for hashingTF.numFeatures and 2 values for lr.regParam,
// this grid will have 3 x 2 = 6 parameter settings for CrossValidator to choose from.
ParamMap[] paramGrid = new ParamGridBuilder()
.addGrid(hashingTF.numFeatures(), new int[]{10, 100, 1000})
.addGrid(lr.regParam(), new double[]{0.1, 0.01})
.build();
// We now treat the Pipeline as an Estimator, wrapping it in a CrossValidator instance.
// This will allow us to jointly choose parameters for all Pipeline stages.
// A CrossValidator requires an Estimator, a set of Estimator ParamMaps, and an Evaluator.
// Note that the evaluator here is a BinaryClassificationEvaluator and its default metric
// is areaUnderROC.
CrossValidator cv = new CrossValidator()
.setEstimator(pipeline)
.setEvaluator(new BinaryClassificationEvaluator())
.setEstimatorParamMaps(paramGrid)
.setNumFolds(2); // Use 3+ in practice
// Run cross-validation, and choose the best set of parameters.
CrossValidatorModel cvModel = cv.fit(training);
// Prepare test documents, which are unlabeled.
DataFrame test = sqlContext.createDataFrame(Arrays.asList(
new Document(4L, "spark i j k"),
new Document(5L, "l m n"),
new Document(6L, "mapreduce spark"),
new Document(7L, "apache hadoop")
), Document.class);
// Make predictions on test documents. cvModel uses the best model found (lrModel).
DataFrame predictions = cvModel.transform(test);
for (Row r: predictions.select("id", "text", "probability", "prediction").collect()) {
System.out.println("(" + r.get(0) + ", " + r.get(1) + ") --> prob=" + r.get(2)
+ ", prediction=" + r.get(3));
}
{% endhighlight %}
## Example: model selection via train validation split
In addition to `CrossValidator` Spark also offers `TrainValidationSplit` for hyper-parameter tuning.
`TrainValidationSplit` only evaluates each combination of parameters once as opposed to k times in
case of `CrossValidator`. It is therefore less expensive,
but will not produce as reliable results when the training dataset is not sufficiently large.
`TrainValidationSplit` takes an `Estimator`, a set of `ParamMap`s provided in the `estimatorParamMaps` parameter,
and an `Evaluator`.
It begins by splitting the dataset into two parts using `trainRatio` parameter
which are used as separate training and test datasets. For example with `$trainRatio=0.75$` (default),
`TrainValidationSplit` will generate a training and test dataset pair where 75% of the data is used for training and 25% for validation.
Similar to `CrossValidator`, `TrainValidationSplit` also iterates through the set of `ParamMap`s.
For each combination of parameters, it trains the given `Estimator` and evaluates it using the given `Evaluator`.
The `ParamMap` which produces the best evaluation metric is selected as the best option.
`TrainValidationSplit` finally fits the `Estimator` using the best `ParamMap` and the entire dataset.
{% highlight scala %}
import org.apache.spark.ml.evaluation.RegressionEvaluator
import org.apache.spark.ml.regression.LinearRegression
import org.apache.spark.ml.tuning.{ParamGridBuilder, TrainValidationSplit}
// Prepare training and test data.
val data = sqlContext.read.format("libsvm").load("data/mllib/sample_libsvm_data.txt")
val Array(training, test) = data.randomSplit(Array(0.9, 0.1), seed = 12345)
val lr = new LinearRegression()
// We use a ParamGridBuilder to construct a grid of parameters to search over.
// TrainValidationSplit will try all combinations of values and determine best model using
// the evaluator.
val paramGrid = new ParamGridBuilder()
.addGrid(lr.regParam, Array(0.1, 0.01))
.addGrid(lr.fitIntercept)
.addGrid(lr.elasticNetParam, Array(0.0, 0.5, 1.0))
.build()
// In this case the estimator is simply the linear regression.
// A TrainValidationSplit requires an Estimator, a set of Estimator ParamMaps, and an Evaluator.
val trainValidationSplit = new TrainValidationSplit()
.setEstimator(lr)
.setEvaluator(new RegressionEvaluator)
.setEstimatorParamMaps(paramGrid)
// 80% of the data will be used for training and the remaining 20% for validation.
.setTrainRatio(0.8)
// Run train validation split, and choose the best set of parameters.
val model = trainValidationSplit.fit(training)
// Make predictions on test data. model is the model with combination of parameters
// that performed best.
model.transform(test)
.select("features", "label", "prediction")
.show()
{% endhighlight %}
{% highlight java %}
import org.apache.spark.ml.evaluation.RegressionEvaluator;
import org.apache.spark.ml.param.ParamMap;
import org.apache.spark.ml.regression.LinearRegression;
import org.apache.spark.ml.tuning.*;
import org.apache.spark.sql.DataFrame;
DataFrame data = jsql.read().format("libsvm").load("data/mllib/sample_libsvm_data.txt");
// Prepare training and test data.
DataFrame[] splits = data.randomSplit(new double[] {0.9, 0.1}, 12345);
DataFrame training = splits[0];
DataFrame test = splits[1];
LinearRegression lr = new LinearRegression();
// We use a ParamGridBuilder to construct a grid of parameters to search over.
// TrainValidationSplit will try all combinations of values and determine best model using
// the evaluator.
ParamMap[] paramGrid = new ParamGridBuilder()
.addGrid(lr.regParam(), new double[] {0.1, 0.01})
.addGrid(lr.fitIntercept())
.addGrid(lr.elasticNetParam(), new double[] {0.0, 0.5, 1.0})
.build();
// In this case the estimator is simply the linear regression.
// A TrainValidationSplit requires an Estimator, a set of Estimator ParamMaps, and an Evaluator.
TrainValidationSplit trainValidationSplit = new TrainValidationSplit()
.setEstimator(lr)
.setEvaluator(new RegressionEvaluator())
.setEstimatorParamMaps(paramGrid)
.setTrainRatio(0.8); // 80% for training and the remaining 20% for validation
// Run train validation split, and choose the best set of parameters.
TrainValidationSplitModel model = trainValidationSplit.fit(training);
// Make predictions on test data. model is the model with combination of parameters
// that performed best.
model.transform(test)
.select("features", "label", "prediction")
.show();
{% endhighlight %}


